Spatiotemporal Glioblastoma Evolution Visual Prediction
Research at The ∀ Lab & Image Science Lab
project status: currently paused (January - September 2025) —
Conducted during the Spring and Summer semesters of 2025 under Dr. Pulkit Grover, Dr. Aswin Sankaranarayanan and Dr. Matthew J Shepard, MD, this study is complementary to a Partial Differential Equation based approach by Cynthia Han a fellow graduate student researcher. Both approaches use a multi-modal approach that combine FLAIR, T1, T2 and CT1 modalities of brain MRI from the LUMIERE dataset.
I worked on a Neural Ordinary Differential (N-ODE) network framework designed to model and predict tumor growth dynamics from longitudinal MRI data. By encoding a patient’s tumor size measurements over time into interpretable kinetic parameters, the N-ODE formulates tumor progression as a continuous-time dynamical system, allowing for unbiased and personalized predictions, even from early-stage data.
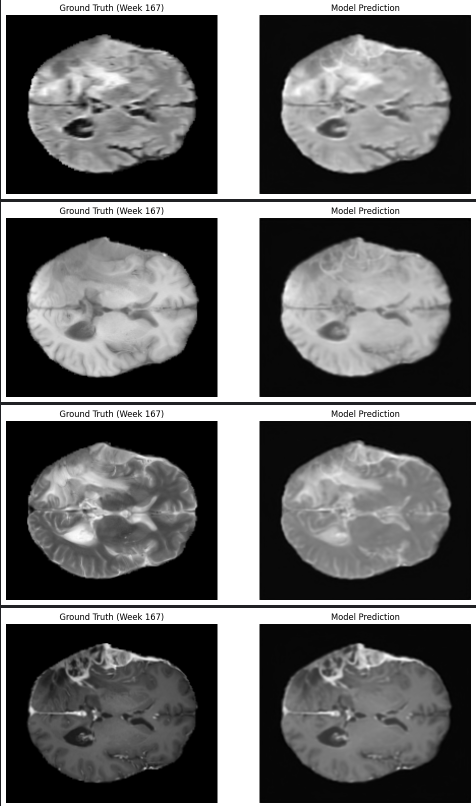
- the model uses a encoder-neural ordinary DE-decoder model with a combined loss approach, integrating Mean Squared Error to match anatomical details and Dice Loss to ensure accurate tumor segmentation, thus optimizing both image fidelity.
- the model’s encoder-decoder architecture not only improves future tumor size predictions but also produces metrics that strongly predict overall survival.